Female Butterflies See UV Light Thanks to a Gene Hiding in an Unusual Place
In some species, female and male butterflies apparently perceive colors differently.
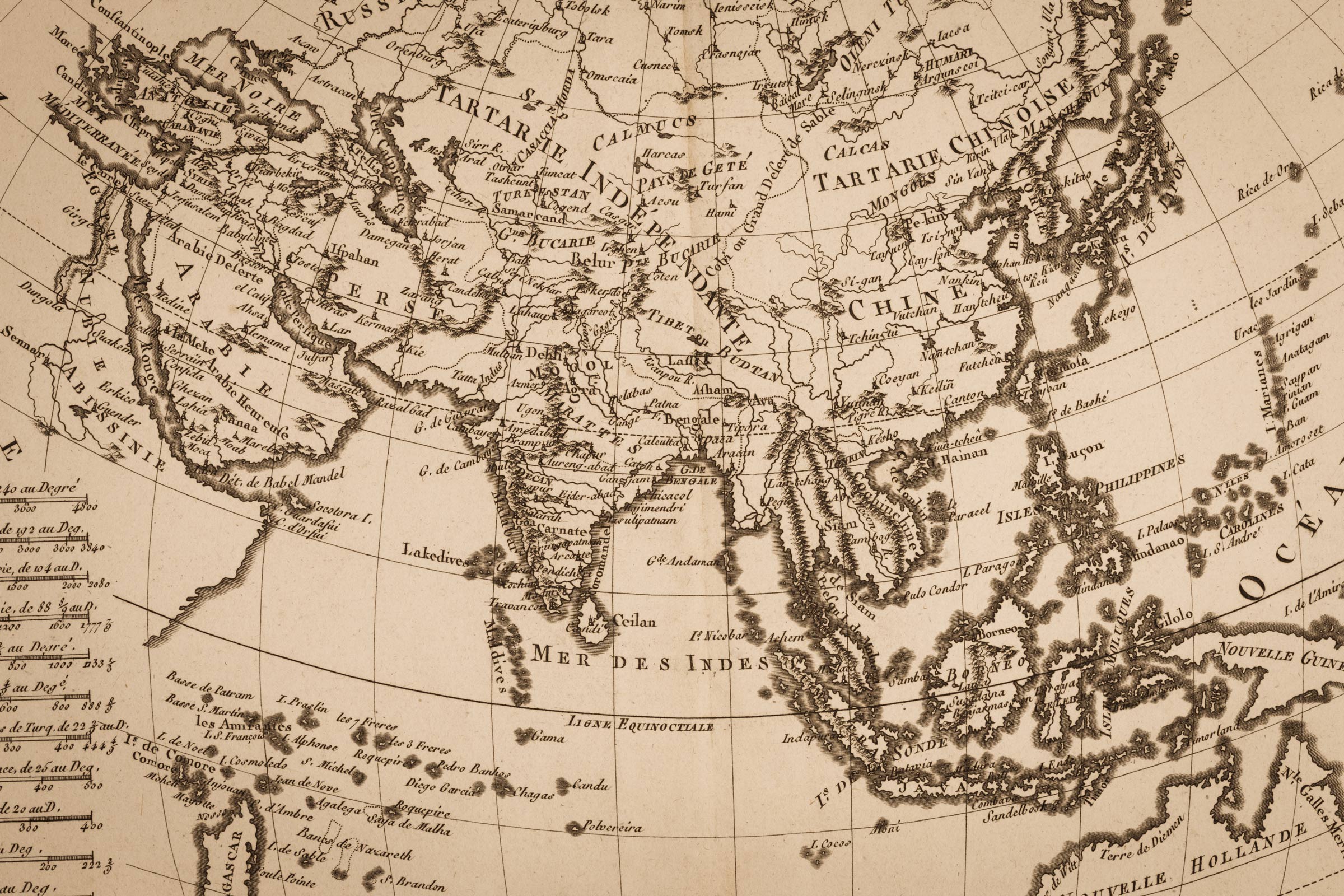
Zebra Longwing (Heliconius charithonia) at the National Butterfly Center in Mission, Texas. Public domain image produced as part of the “Insects Unlocked” project. Photo credit: Alejandro Santillana/University of Texas at Austin.
As butterflies flit among flowers, they don’t all see blossoms the same way. In a phenomenon called sexually dimorphic vision, females of some butterfly species perceive ultraviolet color while the males see light and dark. A team of University of California, Irvine, Texas A&M and University of Texas at Austin biologists, including the Department of Integrative Biology’s Larry Gilbert, has discovered that in at least one species, the variation results from a vision gene’s jump onto a sex chromosome. Theirs is the first known finding that this kind of genetic change causes sexually dimorphic vision.
The study appears in Proceedings of the National Academy of Sciences.
Some Heliconius butterfly species see ultraviolet color, an array wider than the visible light spectrum that humans perceive. A substance produced by the opsin gene accounts for these butterflies’ visual capacity. In one clade of Heliconius species with sexually dimorphic vision, only females see ultraviolet colors.
In searching for the genetic mechanism behind this difference, the researchers selected as their subject Heliconius charithonia, or Zebra Longwing butterfly, whose visual capacity is sexually dimorphic. When the biologists finished assembling the first complete genome for this species, they discovered its W – or female – chromosome contained the opsin gene.
Scientists believe the vision difference may be the reason that females and males within some butterfly species feed on different types of flowers. So far, the only other animals known to have sexually dimorphic vision are some kinds of primates with red-green polymorphism such as humans and squirrel monkeys.
Gilbert said there’s another twist to this story. Adult males of some species of Heliconius will wait for the moment a female emerges from her chrysalis and then immediately attempt to mate with her, as a way to outcompete other males for the chance. This strategy is called pupal mating.
“For now, we are faced with an additional intriguing mystery, as only Heliconius species with males that locate and sit on female pupae, in other words engage in pupal mating, have opsin genes moved to the W chromosome,” Gilbert said.

Two Heliconius charithonia males sit on the pupal case of a female, competing to mate with her when she emerges. Photo credit: Larry Gilbert/University of Texas at Austin.
The revelation about the role of the W chromosome was especially surprising due to the technical difficulty required to recover W chromosomes and because the opsin gene, which plays a vital role in visual perception of light and color, was found in a part of the genome not usually associated with vision.
“This is the first known instance where dimorphic color vision in animals comes from a single gene moving to a sex chromosome,” said first author Mahul Chakraborty, an assistant project scientist in ecology and evolutionary biology at Texas A&M University. “Besides the discovery’s scientific significance, it highlights the complexities of automated genetic sequencing and the crucial role of validation.”
Chakraborty performed much of his work on the project while a postdoctoral researcher in the laboratories of co-corresponding authors Adriana Briscoe and J.J. Emerson at UC Irvine. Gilbert is a professor of integrative biology at UT Austin and director of the Brackenridge Field Laboratory, where Zebra Longwings occur in the wild and also are maintained by Gilbert in a tropical greenhouse for such research at UT Austin and elsewhere.
Previously assembled genomes for Heliconius charithonia were fragmentary. None included the W chromosome, whose highly repetitive code can pose stumbling blocks for automatic sequencing. When the UCI researchers began their work by automatically sequencing the species genome, it failed to reveal all expected copies of the opsin gene. Undeterred, they next examined the coding manually.
“I went through every bit of the sequencing,” said Angelica Lara, who was an ecology and evolutionary biology undergraduate when she began working with the investigative team. She continued to participate in the project as a post-baccalaureate researcher after receiving her degree. “I still couldn’t find the opsin after all that review. Then I realized a part of the code for the W chromosome had not been well-formatted, and I believed the opsin had to be located there.”
Lara’s finding cued Chakraborty to examine that segment more closely. It turned out the automatic sequencing had dropped that section of the chromosome’s coding, likely stymied by its repetitiveness. Restoring it revealed the opsin gene, and the team confirmed the finding with additional tests.
“Without this manual annotation and investigation, we would have made assumptions that were incorrect and misleading,” said Briscoe, a professor of ecology and evolutionary biology. “Now that we’ve made this discovery, we can dig much deeper into the mechanics behind the dimorphism and understand its purpose.”
The research was supported by the National Science Foundation, National Institutes of Health, the UCI Optical Biology Core Facility and Texas A&M University start-up funding.
Adapted from a press release by the University of California, Irvine.